- Research
- Open access
- Published:
MTAF–DTA: multi-type attention fusion network for drug–target affinity prediction
BMC Bioinformatics volume 25, Article number: 375 (2024)
Abstract
Background
The development of drug–target binding affinity (DTA) prediction tasks significantly drives the drug discovery process forward. Leveraging the rapid advancement of artificial intelligence, DTA prediction tasks have undergone a transformative shift from wet lab experimentation to machine learning-based prediction. This transition enables a more expedient exploration of potential interactions between drugs and targets, leading to substantial savings in time and funding resources. However, existing methods still face several challenges, such as drug information loss, lack of calculation of the contribution of each modality, and lack of simulation regarding the drug–target binding mechanisms.
Results
We propose MTAF–DTA, a method for drug–target binding affinity prediction to solve the above problems. The drug representation module extracts three modalities of features from drugs and uses an attention mechanism to update their respective contribution weights. Additionally, we design a Spiral-Attention Block (SAB) as drug–target feature fusion module based on multi-type attention mechanisms, facilitating a triple fusion process between them. The SAB, to some extent, simulates the interactions between drugs and targets, thereby enabling outstanding performance in the DTA task. Our regression task on the Davis and KIBA datasets demonstrates the predictive capability of MTAF–DTA, with CI and MSE metrics showing respective improvements of 1.1% and 9.2% over the state-of-the-art (SOTA) method in the novel target settings. Furthermore, downstream tasks further validate MTAF–DTA’s superiority in DTA prediction.
Conclusions
Experimental results and case study demonstrate the superior performance of our approach in DTA prediction tasks, showing its potential in practical applications such as drug discovery and disease treatment.
Background
The binding of drugs to targets constitutes the foundation for achieving therapeutic efficacy [1], determining whether drugs can effectively modulate physiological functions to treat diseases. A thorough understanding of the interaction mechanisms between drugs and targets is a challenging issue crucial for drug design and development [2]. The strength of interaction between drugs and targets determines the effectiveness of drugs, which can be assessed by measuring binding affinity. This is typically measured by dissociation constant \((K_d)\), inhibition constant \((K_i)\), or half-maximal inhibitory concentration \((IC_{50})\) [3]. Utilizing experimentally determined drug–target binding affinity data to learn the interaction mechanisms between drugs and targets can guide the development of drug discovery. The most dependable approach to ascertain binding affinity is through wet lab experimental measurements. Within the realm of biology, two experimental methodologies employed for quantifying binding affinity are protein microarray technology [4] and affinity chromatography [5]. To delineate effective and safe drugs targeting specific proteins, thousands of compounds must undergo testing [6]. It is usually carried out through high-throughput screening in vitro experiments [7]. Due to the vast number of drug compounds [8] and protein targets [9], some with unknown structures [10], and the dynamic conformational changes upon their binding [11, 12], wet-lab experiments are time-consuming and resource-intensive.
With the advancement of computer technology, researchers have begun to employ computers to predict the interaction between drugs and targets, utilizing docking methods [13] to simulate the binding process of drugs and targets. Computational drug–target affinity (DTA) prediction methods can be categorized into structure-based methods, ligand-based methods, and hybrid methods [14]. AI has become increasingly popular in predicting DTA. In recent years, researchers have developed various AI methods [15] for DTA prediction. These methods can handle large amounts of biological and chemical data, automatically learning useful features to enhance prediction accuracy and efficiency. GraphDTA [16] introduced the paradigm of representing drugs as graphs and leveraged Graph Neural Networks (GNNs) to predict DTA. Comparative analysis against conventional methodologies and alternative deep learning approaches reveals the superior predictive efficacy of GNNs. MSGNN-DTA [17] leverages a fused multi-scale topological feature approach based on GNNs, incorporating a gated skip-connection mechanism during feature learning to fuse multi-scale topological features, thereby yielding information-rich representations of drugs and proteins. This method entails the construction of drug atom graphs, motif graphs, and weighted protein graphs to comprehensively extract topological information.
Since its inception, transformer [18] has been widely applied in the field of deep learning. Its ability to capture latent relationships between sequences has rendered it remarkably effective in DTA prediction tasks. TransformerCPI [19], a novel transformer neural network, introduced a more rigorous label reversal experiment. It can be deconvolved to highlight important interacting regions of protein sequences and compound atoms. The attention mechanism demonstrated its immense potential in predictive tasks within HyperAttentionDTI [20]. It proposed an end-to-end bio-inspired model based on the convolutional neural network (CNN) and attention mechanism. Deep CNNs were used to learn the feature matrices of drugs and proteins. To model complex non-covalent inter-molecular interactions among atoms and amino acids, this study utilized the attention mechanism on the feature matrices and assigned an attention vector to each atom or amino acid.
Although current DTA prediction methods have their respective advantages, many still face several challenges, including: (i) Existing methods mostly focus on the information from molecular graphs and Morgan fingerprints, disregarding the rich information concealed in other modalities. As a result, they often fail to acquire the deep chemical semantic features of drug molecules [21]. Morgan fingerprints primarily concentrate on the local environments within a certain radius of drug molecules [21] (the atomic number, the degree of the atom, the formal charge, and the chemical bonds attached to the atom), overlooking the nuanced features of substructures within other ranges. (ii) Moreover, different features of drugs contribute differently to DTA prediction tasks, and many existing linear fusion methods may not fully exploit the potential of features from each modality [22]. (iii) In fact, during the binding process, the conformations of both drugs molecules [11] and targets [12] continually change, which means that the interaction process between them during fusion is highly complex. However, many current methods utilize interaction modules that are too simplistic to fully capture the mutual interactions between the two [23].
To mitigate these limitations, we propose a method called MTAF–DTA, which is a nested fusion network based on multi-type attention mechanisms, and it utilizes multi-modal features of drugs for DTA prediction. We extracted the Avalon fingerprint, Morgan fingerprint, and molecular graph features of drugs to enrich drug feature information. In drug-related tasks, the combined use of Morgan molecular fingerprints and Avalon molecular fingerprints often demonstrates greater efficacy than their individual use [24], and we get 200 molecular properties, e.g. number of rings, molecular weight, etc. [24]. For the three types of drug information obtained, we designed a drug feature fusion module based on the attention mechanism to update their respective contribution weights. Then we obtain the final drug representation, thus enhancing the capability to capture relevant information regarding drug efficacy. We designed an interaction block SAB, and it involves three times fusion operations utilizing attention mechanisms to integrate the interactions of drug and target information. MTAF–DTA has shown commendable performance on benchmark datasets, comparable to or even surpassing other baseline models. In particular, it attained the best CI and MSE score on the Davis dataset under the Novel-protein data partitioning scheme, outperforming the SOTA method by 1.1% and 9.2%, respectively. This demonstrates its effectiveness in enhancing drug discovery for novel targets. Simultaneously, ablation experiments confirm the effectiveness of each component.
In summary, the main contributions of this study are summarized as follows:
-
We introduced the Avalon molecular fingerprint and integrated it with Morgan fingerprints and molecular graph to capture overlooked chemical semantic information. To the best of our knowledge, MTAF–DTA is the first method to use an attention mechanism to map useful drug features from different modalities into the uniform representation space, thus generating an informative drug representation.
-
To further capture the interactions between drugs and targets, we designed an interaction module named as Spiral-Attention Block (SAB), based on multi-type attention mechanisms. This module better simulates the interaction processes between drugs and target proteins.
-
Our results show the superior predictive capability of MTAF–DTA, with CI and MSE metrics showing respective improvements of 1.1% and 9.2% over the SOTA method in the Davis novel target settings.
Methods
We frame the prediction of DTA as a regression task, utilizing drug simplified molecular input line entry system (SMILES) sequences \(S_d\) and protein sequences \(S_t\) as the input to predict the affinity score for a given drug–target pair. This work aims to determine the binding strength between drugs and target proteins, thereby assisting practical tasks such as drug discovery. Figure 1 shows the overall framework of MTAF–DTA, which consists of four main parts: drug representation module, protein representation module, drug–target feature fusion module, and prediction module.
In the drug representation module, we extract and process three types of features for the drugs and perform feature fusion based on an attention mechanism. Within the protein representation module, an embedding layer maps the amino acid sequences into representation matrices, and then we utilize CNN for feature extraction. These representations of drug and target are then fed into the SAB part, which serves as the drug–target feature fusion module. It is employed to simulate the drug–target interaction process and depict the interaction information between them. By inputting the interaction information into the prediction module, the affinity scores are derived. Additionally, we conducted preprocessing of the dataset before training, as mentioned in Section 3.1.
Drug representation module
Drug feature extraction
To address the issue of information loss in drug molecules and enhance the feature representation capability of drugs, we extracted and integrated multiple drug features to obtain a more comprehensive representation of drugs. Whereas the majority of current DTA methods utilize either molecular graph information or integrate it with Morgan fingerprint [also called extended-connectivity fingerprint(ECFP)] [25] as drug representations, we further augmented this by incorporating Avalon fingerprint [26] features. Ablation experiments demonstrate that this addition contributes to enhancing the predictive accuracy of the model.
-
The Avalon fingerprint is derived from the Avalon Toolkit [27]. It is characterized by its ability to capture geometric and directional information within molecules, which is crucial for depicting molecular shape and spatial conformation.
-
The Morgan fingerprint, constructed based on the Morgan algorithm [28], generates bit-vector representations by considering the local environment of molecules, which are used to describe molecular structure and similarity. The radius parameter determines the range of atomic neighbors considered when constructing the fingerprints.
-
The drug’s SMILES input is preprocessed using the RDkit tool [29] to generate a graph with node features and an adjacency matrix. Then the molecular graph feature is extracted by GNN and utilizes residual connections [30, 31] between layers to prevent information loss. The features of every vertices are acquired through iterative aggregation and propagation of features from neighboring vertices. Eq. (1) shows the message-passing phase:
$$\begin{aligned} y_i^{(k+1)}=F_{k}\left( (1+\omega ^{(k)})y_i^{(k)}+\sum \limits _{j\epsilon N(i)}y_j^{(k)}\right) \end{aligned}$$(1)where \(y_i^{(k)}\epsilon R^m\) represents the feature vector of vertex i at time step k, \(F_k\) is a vertex update function, N(i) is the set of neighboring vertices of vertex i, and \(\omega\) is a learnable parameter. The feature of vertex i updates at the next time step. The entire graph’s feature vector is calculated as Eq. (2).
$$\begin{aligned} F_G=\frac{1}{|V|}\sum \limits _{v\epsilon V}y_v^{(k)} \end{aligned}$$(2)where V represents the set of vertices in the graph.
Inspired by previous work [31], We implemented a technique of random subgraph removal for data augmentation. We randomly select an atom in the drug molecular graph as the initial node. Subsequently, this node is removed, along with its neighboring nodes recursively, until a predetermined proportion of the subgraph is eliminated (0.2 for the Davis dataset and 0.1 for the KIBA dataset) while maintaining the affinity scores of drug–target pairs and the Morgan and Avalon fingerprints of drug molecules unchanged. During the training phase, multiple new pairs are generated, whereas no subgraph removal operation is conducted during the testing phase.
Drug feature fusion module
The information extracted from drug molecules, including Avalon fingerprint \(F_A\epsilon R^d\), Morgan fingerprint \(F_M\epsilon R^d\), and molecular graph information \(F_G\epsilon R^d\), contains different information in DTA prediction tasks. To compute the contribution weights of different drug features and fuse them, we designed a feature fusion module based on the attention mechanism. We first integrated \(F_G\) and \(F_M\), then subsequently incorporated \(F_A\) into the fusion result of the two. This module generates the final drug representation that integrates the effective information from these three types of features.
-
We fused \(F_G\) and \(F_M\) initially. They are represented in the same dimension (denoted as d). The attention weight \(W_{MG}\) can be calculated as follows:
$$\begin{aligned} \begin{aligned} W_{MG}&=Fusion\,Block(F_M+F_G)\\&=\sigma \circ B\circ \tau \circ \delta \circ B\circ \tau (F_M+F_G)\ \end{aligned} \end{aligned}$$(3)where \(\tau\) denotes a linear layer, B represents the Batch Normalization, \(\delta\) denotes the Rectified Linear Unit (ReLU), and \(\sigma\) represents the sigmoid function. \(\circ\) takes the output of the right function as the input of the left function, which means operations in the formula are executed from right to left sequentially. Then, the first fusion can be computed as follows:
$$\begin{aligned} F_{MG}=F_G\cdot W_{MG}+F_M\cdot (1-W_{MG}) \end{aligned}$$(4)
To fully comprehend and efficiently process information about drugs, we designed a model equipped with the ability to learn and integrate multi-modal features. Finally, we incorporate \(F_A\) into this experiment to achieve complementarity between various heterogeneous information.
-
We calculate new fusion weights \(W_{AMG}\) according to the above process using \(F_{MG}\) as the new input, and generate a new output after fusion:
$$\begin{aligned} & W_{AMG}=Fusion\,Block(F_{MG})\end{aligned}$$(5)$$\begin{aligned} & F_{AMG}=F_{MG}\cdot W_{AMG}+F_A\cdot (1-W_{AMG}) \end{aligned}$$(6)The output \(F_{AMG}\) after two rounds of fusion serves as the final output of the drug representation module, and will participate in the subsequent fusion operation with protein representations.
Compared to other methods, We further address the information loss problem of drugs. Mapping useful drug features from different modalities into the uniform representation space allows us to generate an informative drug representation, thereby further enhancing the accuracy of DTA prediction.
Protein representation module
Proteins are represented by amino acid sequences, with each amino acid being denoted by a single uppercase letter, forming a string of uppercase letters. To represent them in a form that deep learning models can process, integer variables are used to encode different amino acids for feature representation. We define this transformation as follows: Let \(convert=(a\rightarrow i; a\epsilon A, i\epsilon I)\), where A is the set of amino acids, for example, “F” represents phenylalanine. And I is an integer set ranging from 1 to 25. Each amino acid sequence is mapped to an integer sequence of uniform length up to 1200. Sequences exceeding this length are truncated, whereas those shorter are padded with zeros. Subsequently, an embedding layer maps each integer in the sequence to a 128-dimensional vector, as shown in Fig. 2.
This yields the representation matrix \(M_{t} \epsilon R^{N\times E}\) of the amino acid sequence, where N represents the maximum length of the amino acid sequence, and E represents the embedding size of the amino acid. It is then passed into the protein representation module, which consists of three layers of 1D-CNN. The feature processing performed at each layer can be represented as:
where \(R_t^{(i)}\) is the i-th hidden protein representation. \(R_t^{(0)}=M_t\), and C denotes a CNN layer.
Drug–target feature fusion module
To better capture the interaction between drug molecules and target proteins, and to minimize errors resulting from insufficient fusion, we devised a drug–target feature fusion module named SAB. It is based on attention mechanisms, which enables the model to concentrate on the most informative features in the interaction process between drugs and targets. The SAB consists of three components:
-
First, the drug feature \(F_D\) and protein feature \(F_T\) are inputted into the Fusion Module to get the fusion feature \(F_{DT}\). The formulas are as follows:
$$\begin{aligned} & W_{1}=Fusion\,Block(F_D+F_T) \end{aligned}$$(8)$$\begin{aligned} & F_{fusion1}=F_{D}\cdot W_{1}+F_T\cdot (1-W_{1}) \end{aligned}$$(9)$$\begin{aligned} & W_{2}=Fusion\,Block(F_{fusion1}) \end{aligned}$$(10)$$\begin{aligned} & F_{DT}=F_{D}\cdot W_{2}+F_T\cdot (1-W_{2}) \end{aligned}$$(11)
-
Next, the drug feature \(F_D\) and protein feature \(F_T\) are passed into a cross-attention module. In the cross-attention mechanism, for each input sequence, similarity scores with other sequences are calculated, and these scores are used to weightedly average the representations of the other sequences, thereby obtaining a cross-modal context representation \(F_{cross1}\) for the current sequence:
$$\begin{aligned} & Q=F_T\end{aligned}$$(12)$$\begin{aligned} & K=V=F_D\end{aligned}$$(13)$$\begin{aligned} & attention\_score=S\left( \frac{QK^{T}}{\sqrt{C/d}}\right) \end{aligned}$$(14)$$\begin{aligned} & \begin{aligned} F_{cross}&=Cross\,Attention(Q,K,V)\\&=attention\_score\cdot V\ \end{aligned} \end{aligned}$$(15)where S in Eq. (14) is a Softmax function outputting attention weights. C and d are the embedding dimensions and number of heads, respectively. The obtained output is added to the drug representation and then passed through a self-attention block, then we get \(F_{self}\):
$$\begin{aligned} & Q=K=V=F_{cross}+F_D\end{aligned}$$(16)$$\begin{aligned} & \begin{aligned} F_{self}&=Self\,Attention(Q,K,V)\\&=attention\_score\cdot V\ \end{aligned} \end{aligned}$$(17)
-
Finally, the two aforementioned outputs are reintroduced into the cross-attention mechanism module for final fusion. The operation can be represented as follows, which represents the final fusion result of the drug–target features:
$$\begin{aligned} F_{d\Leftrightarrow t}=Cross\,Attention(F_{self},F_{DT},F_{DT})\end{aligned}$$(18)
As shown in Fig. 3, the self-attention mechanism differs from the cross-attention mechanism in that it only takes a single-source input.
In conclusion, the comprehensive fusion of drug–target features in our experiment partially simulates the process of drug–target binding, thereby enhancing the prediction accuracy of the model.
Drug–target affinity prediction
The fusion result of drug and target features \(F_{d\Leftrightarrow t}\) leads to the following equation for the prediction process:
\({\hat{y}}\) represents the predicted affinity score of the drug–target pair. “3” denotes that the process within the parentheses is repeated three times.
The overall DTA prediction model is trained by minimizing the following Mean Squared Error (MSE) loss function, which is used to quantify the disparity between predicted values and true values:
where \(y_i\) is the true affinity score of i-th drug–target pair, and N is the sample size. A smaller MSE indicates a closer alignment between predicted and true values, which is indicative of enhanced model performance.
Algorithm 1 provides a detailed description of the algorithm for training the proposed MTAF–DTA model.
Results
Dataset
We benchmarked performance on two commonly used datasets: Davis [32] and KIBA [33]. Additionally, we expanded our investigation by model downstream studies on four datasets, including BindingDB [34] and Metz [35], in addition to the aforementioned datasets. Furthermore, we conducted a case study on the DrugBank [36]. Detailed information regarding each task will be presented in their respective sections. The detailed information of Davis and KIBA utilized in the regression tasks is as follows:
-
The Davis dataset, created by Davis et al. comprises 68 compounds along with their binding affinity data for 442 protein targets. Each compound has experimentally determined dissociation constant \((K_d)\) values with their respective targets. This value reflects the strength of binding between the drug molecule and the target protein. The \(K_d\) values obtained in the experiment are transformed as follow:
$$\begin{aligned} pK_d=-\log _{10}(K_d/10^9)\end{aligned}$$(21)
-
Tang et al. introduced a model-based integration approach called KIBA to generate an integrated drug–target bioactivity matrix. The KIBA dataset comprises approximately 2111 compounds along with binding affinity data for 229 targets. It uses the KIBA score to represent the interaction between the drug and the target protein.
We discovered that the datasets suffer from data duplication problems, wherein identical drug–target pairs are associated with different affinity scores. It may affect the training process, thereby reducing the predictive performance of the model. Table 1 shows the statistical information of the two datasets.
Redundancy represents the quantity of duplicated drug–target pairs within the dataset, and the \(Redundancy\ rate\) is computed as follow:
where N represents the total number of drug–target pairs of the dataset.
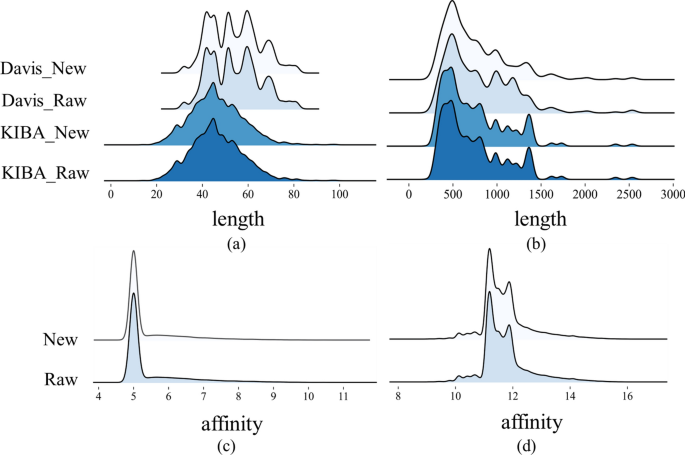
This study implemented distinct preprocessing strategies for the two datasets based on the proportion of duplicated drug–target pairs within the entire dataset. As for Davis, we averaged the affinity scores of all duplicate drug–target pairs to obtain the final value used in training. For the KIBA dataset, we removed all duplicate pairs. The Davis dataset exhibits a high redundancy rate of 18.3\(\%\), thus implementing a deduplication measure would impact the overall distribution of the dataset. As shown in Fig. 4, our operations do not change the overall distribution of the datasets.
Randomly splitting datasets (where drugs and targets in the test set have already appeared in the training set) may cause information leakage and make the results overly optimistic [37]. From an application perspective, most proteins or drugs do not appear in the training [38]. In this study, we followed the given methodology to divide the dataset, which was implemented by the open-source software DeepPurpose [39]:
-
Novel-protein: There is no overlap between the proteins in the test set and those in the training set. Additionally, all drugs will be present in both sets.
-
Novel-drug: The drugs used in the test set do not overlap with those used in the training set, whereas all proteins are present in both the test and training sets.
-
Novel-pair: There is no intersection between drugs and proteins in either the test or training sets.
Each of the three data partitioning methods above addresses different research objectives. The model trained using the Novel-protein partitioning approach facilitates drug discovery for novel proteins. Models trained using the Novel-drug partitioning method are useful for identifying interacting proteins for newly developed drug compounds. The Novel-pair approach provides valuable information on the binding of novel proteins to newly developed drugs.
Performance evaluation metrics
To evaluate the performance of various models, our study employed regression evaluation metrics MSE loss, and the Concordance Index (CI) [40]. CI is defined as the proportion of label pairs for which the predicted outcome is consistent with the actual outcome. The formula for CI is as follows:
where \(d_i\) and \(d_j\) are distinct true label values, with \(d_i>d_j\), and \(b_i\) and \(b_j\) are the corresponding predicted values. If the relative ordering of two predicted values matches that of the true values, the indicator function h(x) returns a value of 1. It returns 0.5 or 0 if they are equal or opposite. In Eq. (23), Z represents the number of ordered pairs of affinity label values in the dataset. CI value ranges from 0 to 1. A CI of 1 indicates that all ordering of affinity scores is correctly predicted, whereas a CI of 0 signifies that all ordering is incorrectly predicted. Higher CI values imply stronger predictive capability.
Settings of hyperparameters and experimental environment
All experiments in this study were conducted on four NVIDIA RTX A6000 (48G) GPUs. The implementation was carried out using Python version 3.8.0 (default, Nov 6, 2019, 21:49:08) and PyTorch [41] version 2.0.1, with training facilitated by the Adam optimizer [42]. Additionally, we employed the Cosine Annealing scheduler (CosineLRScheduler) encapsulated in the timm library, with parameter configurations detailed in Table 2. Batch sizes for each dataset were determined within the range: [32, 64, 128, 256], with a batch size of 64 set for the Davis dataset and 128 for the KIBA dataset. The training epochs were set to 3000, with early stopping applied at 500 epochs.
Performance comparison with baseline methods
Baseline methods
This section presents a comparative analysis of the experimental results between MTAF–DTA and baseline methods on benchmark datasets Davis and KIBA. To validate the effectiveness of the proposed MTAF–DTA model, we compared it against the following baseline methods: Support Vector Machine (SVM), Random Forest (RF); DeepDTA [3]; TransformerCPI [19]; MGraphDTA [43]; ColdDTA [31]; AttentionMGT-DTA [44].
However, as shown in Table 3, these methods insufficiently extract and fuse drug features, leading to drug information loss, which negatively impacts model prediction outcomes. Furthermore, approaches relying on linear concatenation for drug–target integration lack a simulation of the fusion process between them. In response to these limitations, MTAF–DTA incorporates enhanced drug representation fusion and employs a multi-type attention mechanism for drug–target integration, effectively mitigating some of these issues. In comparison with the method that leverages AlphaFold2 for protein information extraction, MTAF–DTA still demonstrates superior competitiveness. Under the cold-start partitioning of the Davis dataset, our CI and MSE metrics consistently surpass that of AttentionMGT-DTA. Specifically, we expanded the types of drug features to include molecular graph information, Morgan fingerprints, and Avalon fingerprints, thereby extracting a richer set of chemical semantic information compared to existing methods. Furthermore, we employed an attention-based fusion module to map the various modalities of drug features into a uniform representation space, resulting in the final drug representation. This operation assigns different attention weights to the diverse modal features, maximizing the exploitation of their respective contributions.
Performance comparison
The superior performance of MTAF–DTA underscores its effectiveness in the task of DTA prediction. We employed a five-fold cross-validation method, in which all data were evenly partitioned into five parts, with one part used as the test set and the remaining four parts used for training. The results for each evaluation metric were derived as the average of five cross-validation iterations. To ensure fairness, we trained baseline models using the optimal parameter settings determined by the baseline method or directly utilized results reported in their published papers. This approach was adopted to mitigate potential errors that may arise during the experimental execution. However, differences in dataset partitioning randomness and variations in experimental machine performance may lead to discrepancies between our experimental outcomes and those reported in other studies.
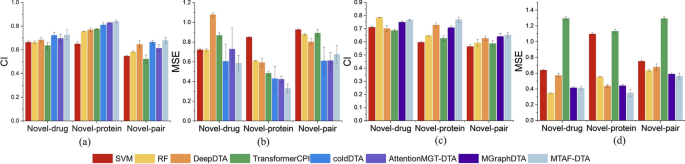
Supplementary Table 1, Additional file 1 illustrates the comparison between different baseline methods and MTAF–DTA on the Davis dataset. The results indicate that MTAF–DTA achieves the best prediction performance in terms of the CI metric across three distinct data partitioning schemes: Novel-drug, Novel-protein, and Novel-pair. Specifically, under the Novel-protein partitioning scheme, MTAF–DTA outperforms the SOTA method by 1.1% in CI and reduces the MSE by 9.2%, yielding values of 0.840 and 0.330, respectively. These results signify that our designed drug feature extraction and fusion modules bolster the representation capability of drug features, effectively enhancing the predictive capacity for the affinity between known drugs and novel protein targets.
On the KIBA dataset, as shown in Supplementary Table 2, Additional file 1, we compared MTAF–DTA with SVM, RF, DeepDTA, TransformerCPI, and MGraphDTA. Traditional machine learning method random forest achieved the best performance on the Novel-drug partitioning, with CI and MSE reaching 0.785 and 0.348, respectively, indicating that machine learning methods still possess competitive capability in DTA prediction tasks [45]. Under this partitioning scheme, MTAF–DTA demonstrated improvements compared to other methods. Specifically, it outperformed SVM by 5.4%, DeepDTA by 6.4%, TransformerCPI by 7.9%, and MGraphDTA by 1.6% in terms of CI. Notably, our proposed method continued to achieve the best performance on the Novel-protein partitioning. Here, the CI reached 0.769, surpassing the values of other methods: SVM (0.596), RF (0.647), DeepDTA (0.728), TransformerCPI (0.627), and MGraphDTA (0.708), with respective improvements of 17.3%, 12.2%, 4.1%, 14.2%, and 6.1% in CI.
To visualize the performance improvement of MTAF–DTA, we plotted the CI and MSE from Supplementary Table 1, Additional file 1 and Supplementary Table 2, Additional file 1, as shown in Fig. 5. It is evident that our method exhibits significant advantages. MTAF–DTA has achieved notable performance improvement on the Novel-protein partitioning, attributed to the extraction and integration of richer drug features. Additionally, the SAB part further enhanced the prediction performance of the model.
Performance of the model on downstream tasks
The insufficient simulation of the drug–target binding process is indeed a challenge faced by current AI methods, and this is one of the primary motivations for our research. Given the extensive exploration of drug features in our method and the demonstrated potential of our research in identifying potentially effective drugs for novel proteins in the regression tasks, this section primarily focuses on experimental validation under the Novel-protein partitioning scheme.
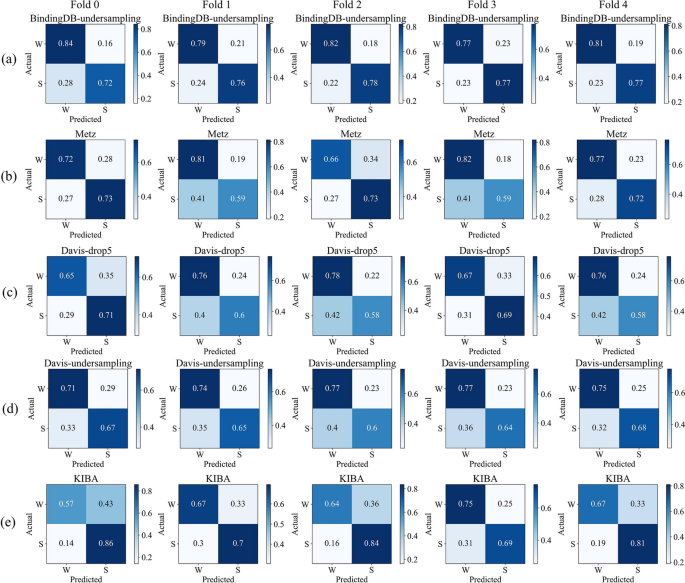
We conducted our study on the BindingDB, Metz, Davis, and KIBA datasets. Considering the different distributions of affinity values in each dataset, we applied different thresholds to classify them into strong and weak bindings. To balance the datasets, we undersampled 16,699 entries from the BindingDB dataset. For the Davis dataset, we either undersampled 1037 drug–target pairs with affinity values of 5 or completely removed them. The statistics of each dataset are presented in Table 4.
We visualized the normalized confusion matrices obtained after MTAF–DTA’s prediction. Figure 6 presents the results of BindingDB-undersampling, Metz, Davis-drop5, Davis-undersampling, and KIBA under five-fold cross-validation. It is evident that the majority of molecules can be accurately predicted across all datasets. And our approach demonstrates a relatively low probability of predicting false positives. For the undersampled BindingDB dataset, our method achieved an average false positive rate of 19.4\(\%\) under five-fold cross-validation, whereas Metz reported 24.4\(\%\) and KIBA 34\(\%\). The balanced Davis dataset, which excluded affinity values of 5, exhibited a false positive rate of 27.6\(\%\), while the undersampled dataset showed a false positive rate of 25.2\(\%\). In conclusion, MTAF–DTA performs well in predicting DTA for potential drugs targeting novel proteins.
Comparison and analysis of data processing
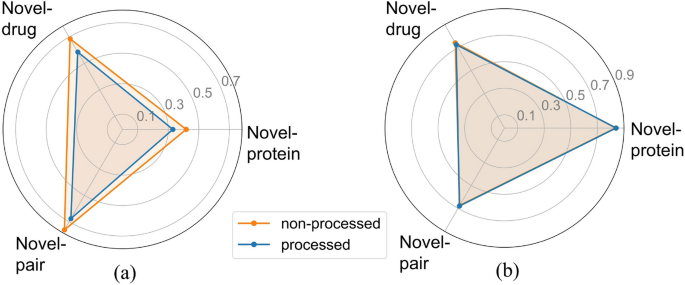
Our experiments, like most studies, were conducted on public datasets. However, unlike previous approaches, we addressed the problem of the dataset containing different affinity scores for the same drug–target pairs. To justify this procedure, we conducted a comparative experiment between the processed and unprocessed datasets. All other hyperparameters and experimental conditions remained consistent throughout the study.
Figure 7a illustrates a significant optimization in MSE following data processing, affirming that the presence of diverse affinity scores indeed influences the model training process, potentially diminishing the predictive capability of the model. The CI metric results in Fig. 7b further validate this observation, showing enhancements in two partitioning schemes for the processed dataset (Novel-protein: 0.840, Novel-drug: 0.726, Novel-pair: 0.679) compared to the unprocessed data (Novel-protein: 0.836, Novel-drug: 0.741, Novel-pair: 0.673). Based on the comprehensive experimental results, we ultimately retained the data preprocessing step. It is noteworthy that the results obtained from training on the raw data still exhibit advantages in comparison to baseline methods, demonstrating the superiority of MTAF–DTA in DTA prediction tasks.
Ablation study
To validate the effectiveness of the components proposed in our method, we conducted the following ablation experiments on the Davis dataset. Specifically, we examined the impact of drug–target fusion methods, drug feature fusion methods, and the quantity and types of drug features on the DTA prediction task. The procedures are as follows:
-
\(AMG-L_{dp}SAB\): Replacing the multi-modal fusion of drug and protein features with a simple linear operation.
-
\(L_{AMG}-SAB\): Replacing the complex fusion method in our approach with a simple linear summation of drug features.
-
\(MG-SAB\): Reducing the fusion of drug features from molecular graph features, Morgan fingerprint, and Avalon fingerprint to a fusion of molecular graph features and Morgan fingerprint, employing the same fusion method as the full MTAF–DTA model which was denoted as baseline.
The experimental results, as depicted in Supplementary Table 3, Additional file 1, demonstrate the effectiveness of the aforementioned modules in the DTA prediction task. Under the Novel-drug partitioning scheme, selecting only Morgan fingerprint and molecular graph features as drug features resulted in a 1.5% decrease in CI. This result validates the effectiveness of our approach in selecting drug features for the DTA prediction task. Within the Novel-protein partitioning scheme, the \(AMG-L_{dp}SAB\) operation led to a 1.3% decrease in the CI metric. In the \(L_{AMG}-SAB\) ablation experiment, only the MSE metric showed a 0.5% improvement under the Novel-pair partitioning scheme, whereas the CI metric decreased by 1.8%, indicating the crucial importance of both drug feature fusion and drug–target feature fusion across the entire DTA prediction task.
Case study
In this section, we present case studies focusing on specific drugs and proteins. We randomly selected two proteins from DrugBank and a subset of drugs associated with each protein, as well as two drugs and a subset of proteins associated with each drug, to constitute our test set.
As shown in Tables 5 and 6, MTAF–DTA achieved a prediction accuracy of 100% for proteins related to Moxisylyte, and the prediction accuracy for Lindane reached 90%. For the proteins lg gamma-1 chain C region and Microtubule-associated protein tau (MAPT), we achieved prediction accuracies of 90% and 80%, respectively. These experimental results effectively demonstrate the capability of MTAF–DTA to screen relevant drugs for specific target proteins and also validate its superior ability in DTA prediction tasks.
Discussion
Despite the significant improvement in predictive performance achieved by MTAF–DTA, it still possesses certain limitations. The improvement in predictive performance of our method on Novel-drug and Novel-pair partitioning is not as pronounced as that on Novel-protein partitioning, suggesting a substantial room for improvement in identifying potential proteins that may interact with new drugs. This could be attributed to our sufficiently comprehensive extraction of drug information, whereas enhancing the extraction of protein information, such as augmenting the extraction of protein 3D structural information, represents a direction for our future improvements. Indeed, the emergence of novel drugs and proteins does not adhere to specific patterns, presenting a new challenge in accurately predicting DTA with limited data. This also implies that the iterative updates of datasets will accompany the development of DTA prediction tasks. Furthermore, the prediction of DTA may even necessitate the application of few-shot learning methods, such as meta-learning. These aspects warrant further investigation and consideration in future research endeavors.
Conclusion
In this paper, we introduce MTAF–DTA, a model developed in response to the limitations of current machine learning methods used for predicting DTA. MTAF–DTA maps useful drug features from different modalities into the uniform representation space to generate an informative drug representation, addressing the drug information loss problem in current methods. Additionally, the SAB fusion strategy of MTAF–DTA improves upon existing fusion approaches, effectively capturing the complex relationship between drugs and targets. It mitigates the DTA prediction errors caused by conformational changes during their binding process. We conducted DTA prediction under cold start settings, according to practical application scenarios. Experimental results demonstrate that MTAF–DTA achieves SOTA performance in the novel target setting and is competitive or superior to baseline models in both novel drug and novel pair settings. These findings showcase that MTAF–DTA effectively enhances the prediction accuracy of DTA. Extensive downstream tasks and case studies further substantiate the potential of MTAF–DTA in drug discovery tasks, particularly in identifying potential effective drugs for novel disease proteins.
Availability of data and materials
The source codes and datasets are publicly accessible at https://github.com/shly-lab/MTAF-DTA.
Abbreviations
- DTA:
-
Drug–target affinity
- AI:
-
Artificial intelligence
- ECFP:
-
Extended-connectivity fingerprint
- SMILES:
-
Simplified molecular input line entry system
- SAB:
-
Spiral-attention block
- SOTA:
-
State-of-the-art
- GCN:
-
Graph convolutional network
- CNN:
-
Convolutional neural network
- CI:
-
Concordance index
- MSE:
-
Mean squared error
- SVM:
-
Support vector machine
- RF:
-
Random forest
References
Wang H. Prediction of protein–ligand binding affinity via deep learning models. Brief Bioinform. 2024;25(2):081.
Öztürk H, Ozkirimli E, Özgür A. Widedta: prediction of drug–target binding affinity (2019). arXiv preprint arXiv:1902.04166
Öztürk H, Özgür A, Ozkirimli E. Deepdta: deep drug–target binding affinity prediction. Bioinformatics. 2018;34(17):821–9.
Lee H, Lee JW. Target identification for biologically active small molecules using chemical biology approaches. Arch Pharmacal Res. 2016;39:1193–201.
Schirle M, Jenkins JL. Identifying compound efficacy targets in phenotypic drug discovery. Drug Discov Today. 2016;21(1):82–9.
Peng J, Wang Y, Guan J, Li J, Han R, Hao J, Wei Z, Shang X. An end-to-end heterogeneous graph representation learning-based framework for drug–target interaction prediction. Brief Bioinform. 2021;22(5):430.
He H, Chen G, Chen CY-C. Nhgnn-dta: a node-adaptive hybrid graph neural network for interpretable drug–target binding affinity prediction. Bioinformatics. 2023;39(6):355.
Zhang L, Wang C-C, Zhang Y, Chen X. Gpcndta: prediction of drug–target binding affinity through cross-attention networks augmented with graph features and pharmacophores. Comput Biol Med. 2023;166:107512.
Zhu Z, Yao Z, Zheng X, Qi G, Li Y, Mazur N, Gao X, Gong Y, Cong B. Drug–target affinity prediction method based on multi-scale information interaction and graph optimization. Comput Biol Med. 2023;167:107621.
Burley SK, Berman HM, Bhikadiya C, Bi C, Chen L, Di Costanzo L, Christie C, Dalenberg K, Duarte JM, Dutta S, et al. Rcsb protein data bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2019;47(D1):464–74.
Lv Q, Zhou J, Yang Z, He H, Chen CY-C. 3d graph neural network with few-shot learning for predicting drug–drug interactions in scaffold-based cold start scenario. Neural Netw. 2023;165:94–105.
Ayaz P, Lyczek A, Paung Y, Mingione VR, Iacob RE, Waal PW, Engen JR, Seeliger MA, Shan Y, Shaw DE. Structural mechanism of a drug-binding process involving a large conformational change of the protein target. Nat Commun. 2023;14(1):1885.
Rarey M, Kramer B, Lengauer T, Klebe G. A fast flexible docking method using an incremental construction algorithm. J Mol Biol. 1996;261(3):470–89.
Sydow D, Burggraaff L, Szengel A, Vlijmen HW, IJzerman AP, Westen GJ, Volkamer A. Advances and challenges in computational target prediction. J Chem Inf Model. 2019;59(5):1728–42.
Bagherian M, Sabeti E, Wang K, Sartor MA, Nikolovska-Coleska Z, Najarian K. Machine learning approaches and databases for prediction of drug–target interaction: a survey paper. Brief Bioinform. 2021;22(1):247–69.
Nguyen T, Le H, Quinn TP, Nguyen T, Le TD, Venkatesh S. Graphdta: predicting drug–target binding affinity with graph neural networks. Bioinformatics. 2021;37(8):1140–7.
Wang S, Song X, Zhang Y, Zhang K, Liu Y, Ren C, Pang S. Msgnn-dta: multi-scale topological feature fusion based on graph neural networks for drug–target binding affinity prediction. Int J Mol Sci. 2023;24(9):8326.
Ashish V. Attention is all you need. Adv Neural Inf Process Syst. 2017;30:I.
Chen L, Tan X, Wang D, Zhong F, Liu X, Yang T, Luo X, Chen K, Jiang H, Zheng M. Transformercpi: improving compound-protein interaction prediction by sequence-based deep learning with self-attention mechanism and label reversal experiments. Bioinformatics. 2020;36(16):4406–14.
Zhao Q, Zhao H, Zheng K, Wang J. Hyperattentiondti: improving drug–protein interaction prediction by sequence-based deep learning with attention mechanism. Bioinformatics. 2022;38(3):655–62.
Fang S, Liu Y, Liu S. Mfgb: molecular properties prediction leveraging self-supervised morgan fingerprint representation learning. In: 2023 IEEE international conference on bioinformatics and biomedicine (BIBM), IEEE; 2023. pp. 3004–3011
Dehghan A, Abbasi K, Razzaghi P, Banadkuki H, Gharaghani S. Ccl-dti: contributing the contrastive loss in drug–target interaction prediction. BMC Bioinform. 2024;25(1):48.
Jiang M, Li Z, Zhang S, Wang S, Wang X, Yuan Q, Wei Z. Drug–target affinity prediction using graph neural network and contact maps. RSC Adv. 2020;10(35):20701–12.
Notwell JH, Wood MW. Admet property prediction through combinations of molecular fingerprints (2023). arXiv preprint arXiv:2310.00174
Rogers D, Hahn M. Extended-connectivity fingerprints. J Chem Inf Model. 2010;50(5):742–54.
Gedeck P, Rohde B, Bartels C. Qsar- how good is it in practice? Comparison of descriptor sets on an unbiased cross section of corporate data sets. J Chem Inf Model. 2006;46(5):1924–36.
Glodek A, Horrigan S, Wachtel K, Katz S, Glanowski S, Soppet D, Liu D, Hise D, Castaneda J, Ebner R, et al. Avalon compound profiling toolkit: use of gene transcription biomarker signatures for drug discovery. Cancer Res. 2008;68(9-Supplement):796–796.
Morgan HL. The generation of a unique machine description for chemical structures-a technique developed at chemical abstracts service. J Chem Doc. 1965;5(2):107–13.
Landrum G. Rdkit: open-source cheminformatics. 2006. Google Scholar (2006)
He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition (2016), pp. 770–778
Fang K, Zhang Y, Du S, He J. Colddta: utilizing data augmentation and attention-based feature fusion for drug–target binding affinity prediction. Comput Biol Med. 2023;164:107372.
Davis MI, Hunt JP, Herrgard S, Ciceri P, Wodicka LM, Pallares G, Hocker M, Treiber DK, Zarrinkar PP. Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol. 2011;29(11):1046–51.
Tang J, Szwajda A, Shakyawar S, Xu T, Hintsanen P, Wennerberg K, Aittokallio T. Making sense of large-scale kinase inhibitor bioactivity data sets: a comparative and integrative analysis. J Chem Inf Model. 2014;54(3):735–43.
Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK. Bindingdb: a web-accessible database of experimentally determined protein–ligand binding affinities. Nucl Acids Res. 2007;35(suppl–1):198–201.
Metz JT, Johnson EF, Soni NB, Merta PJ, Kifle L, Hajduk PJ. Navigating the kinome. Nat Chem Biol. 2011;7(4):200–2.
Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, Chang Z, Woolsey J. Drugbank: a comprehensive resource for in silico drug discovery and exploration. Nucl Acids Res. 2006;34(suppl-1):668–72.
Liu J, Shen Z, He Y, Zhang X, Xu R, Yu H, Cui P. Towards out-of-distribution generalization: a survey (2021). arXiv preprint arXiv:2108.13624
Bemis GW, Murcko MA. The properties of known drugs. 1. Molecular frameworks. J Med Chem. 1996;39(15):2887–93.
Huang K, Fu T, Glass LM, Zitnik M, Xiao C, Sun J. Deeppurpose: a deep learning library for drug–target interaction prediction. Bioinformatics. 2020;36(22–23):5545–7.
Gönen M, Heller G. Concordance probability and discriminatory power in proportional hazards regression. Biometrika. 2005;92(4):965–70.
Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, Killeen T, Lin Z, Gimelshein N, Antiga L, et al. Pytorch: a imperative style, high-performance deep learning library. Adv Neural Inf Process Syst. 2019; 32
Kingma DP, Ba J. Adam: a method for stochastic optimization (2014). arXiv preprint arXiv:1412.6980
Yang Z, Zhong W, Zhao L, Chen CY-C. Mgraphdta: deep multiscale graph neural network for explainable drug–target binding affinity prediction. Chem Sci. 2022;13(3):816–33.
Wu H, Liu J, Jiang T, Zou Q, Qi S, Cui Z, Tiwari P, Ding Y. Attentionmgt-dta: a multi-modal drug–target affinity prediction using graph transformer and attention mechanism. Neural Netw. 2024;169:623–36.
Bai P, Miljković F, John B, Lu H. Interpretable bilinear attention network with domain adaptation improves drug–target prediction. Nat Mach Intell. 2023;5(2):126–36.
Acknowledgements
Not applicable.
Funding
This work was supported in part by the Ministry of Science and Technology under Grant 2022FY101104, and in part by the Natural Science Foundation of Beijing, China under Grant 7244507.
Author information
Authors and Affiliations
Contributions
J.S. proposed and designed the model concept, conducted experiments, and drafted the manuscript. H.W. provided guidance on the model design and revised the manuscript. J.M. guided the visualization and revised the manuscript. J.W. and J.G. offered insightful guidance on algorithm design, analyzed the results, and contributed to manuscript revision through constructive discussions. All authors made significant contributions to the writing and revision of the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and Consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sun, J., Wang, H., Mi, J. et al. MTAF–DTA: multi-type attention fusion network for drug–target affinity prediction. BMC Bioinformatics 25, 375 (2024). https://doiorg.publicaciones.saludcastillayleon.es/10.1186/s12859-024-05984-3
Received:
Accepted:
Published:
DOI: https://doiorg.publicaciones.saludcastillayleon.es/10.1186/s12859-024-05984-3